Research & Software
Our research covers topics of practical computer science and life science informatics. Many of the developed algorithms and methods are also implemented in our open source software.
| Research Methodologies |
|---|
Network Analysis and Visualisation
At any moment in time, we are driven by and are an integral part of many interconnected, dynamically changing networks. Networks in the life sciences can represent a diversity of information such as metabolism, gene regulation, neuronal connections, spread of infections, and food webs. These networks can be assigned to different (interconnected) levels, may change over time and usually have additional data related to their elements (nodes and edges). We research algorithms and methods for investigating network properties as well as for network visualisation and exploration in order to understand the shape, central elements, motifs etc. of networks.
B. H. Junker and F. Schreiber: ‘Analysis of biological networks’ Wiley Book Series on Bioinformatics, Computational Techniques and Engineering, Wiley, 368 pages, 2008.
Immersive Analytics

We are living in a complex, three-dimensional environment which makes use of all our senses. Immersive analytics investigates how new interaction and display technologies can be used to support a deep cognitive, perceptual, and/or emotional involvement when understanding and reasoning about data. It focuses on multi-sensory interfaces for collaboration and for facilitating the immersion of users in their data, aiming at a real life experience. We do both basic research in immersive analytics (such as granularity of immersion, task performance, collaboration in immersive environments) as well as the development of immersive analytics applications in the life sciences. See also here for more information.
K. Marriott, F. Schreiber et al.: ‘Immersive Analytics’ Springer LNCS 11190, 366 pages, 2018.
Modelling and Simulation
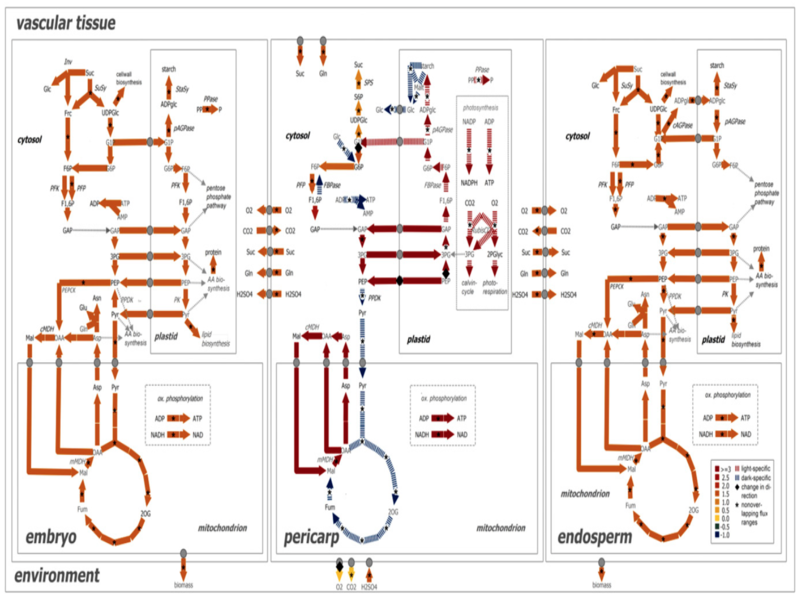
Life is a dynamic process. Computational modeling helps in understanding biological processes (from cellular events to whole ecosystems) as well as in hypothesis generation (such as for drug discovery). We research a range of topics related to modeling and simulation in the life sciences from new modelling and analysis approaches; to information systems representating biological pathways and networks; to modelling, simulation, and evaluation of cellular processes (from metabolism to cellular membranes to cells); to visualisation and immersive analytics methods for exploring data, simulation and analysis results.
I. Koch, W. Reisig and F. Schreiber: ‘Modeling in systems biology: the Petri net approach’ Springer Book Series Computational Biology, Springer, 364 pages, 2011.
Knowledge Representation and Data Integration
Information and knowledge in the life sciences is often distributed and unstructured, and experimental methods result in diverse data sets about the same biological object. Ontologies, standards and methods for their analysis and exploration help in better knowledge representation and reasoning and provide a basis for better data integration. We are part of larger consortia for standard and ontology development and we conduct research on algorithms and methods for working with standards in the life sciences (such as verification and translation of standards and their usage for knowledge discovery).
F. Schreiber, B. Sommer et al.: ‘Specifications of standards in systems and synthetic biology: Status and developments in 2020’ Special issue Journal of Integrative Bioinformatics 17(2-3), 20200022, 2020.
| Research Applications |
|---|
In the following we present some examples for applications of our research methodologies. We are not only interested in theoretical and practical problems from bioinformatics and computer science but also in the underlying biological and medical questions.
Cellular processes in bacteria (Pseudomonas aeruginosa, Acinetobacter baumannii)
Twelve families of bacteria, including Pseudomonas and Acinetobacter, are listed by the WHO as “priority pathogens” that pose the greatest threat to human health. Together with pharmacologists and microbiologists we model parts of these two bacterial systems (e.g., cellular membranes with molecular dynamics, metabolism with constraint-based models) and investigate those models for antibiotics research. The long-term aim is to develop whole cell models based on all relevant sub-systems.
Animal Collective Behaviour (Birds, Fish, Mammals)
We are part of the university’s cluster of excellence “Centre for the Advanced Study of Collective Behaviour” and develop computer science methods for integrated studies of collective behaviour across a wide range of species. We work together with biologists to understand collective behaviour, for example, in bird flocks and fish swarms, and are contributing to the development of the MoveBank as well as novel data analytics approaches for the space-borne animal tracking system ICARUS.
Toxicology (Microcystines, Neurotoxicity)
The toxicity of many substances is not well understood. Together with toxicologists we develop algorithms for the risk assessment of toxins, such as microcystines (which are produced by cyanobacteria species in water bodies worldwide and represent one of the toxin types most frequently associated with drinking water, food supplement and/or food contamination) and chemicals with neurotoxicity potential.
Plant Metabolism (Barley, Maize, Pea, Canola)
Just three of the worlds over 50,000 edible plants – maize, rice and wheat – provide more than 50 percent of the world's food energy intake. Together with plant biologists we collect, structure and analyse data regarding central metabolism in major crop plants (such as maize, barley as model plant for wheat, and canola). We build and analyse models (constraint-based, kinetic models, etc.) including novel multiscale models to capture and investigate the plant’s metabolic processes.
Human Behaviour (Sitting Behaviour, Activity)
“Sitting is the new smoking”, and sitting or lying down for too long is a major contributor to your risk of chronic health problems, such as diabetes, heart disease, and cancer. Together with scientists from medicine, psychology and design we investigate novel ways for personalised interventions to promote health, for example, to improve the sitting behaviour of people.
Ecology and Environment (Great Barrier Reef)
The Great Barrier Reef is considered the largest organism in the world, and it is at critical risk of rapid decline. Together with marine biologists and ecologists we research on integration and visualisation methods for data from multiple resources to provide users insights for education, exploration and decision-making purposes.
Software |
|---|
The following software is open source.
Vanted
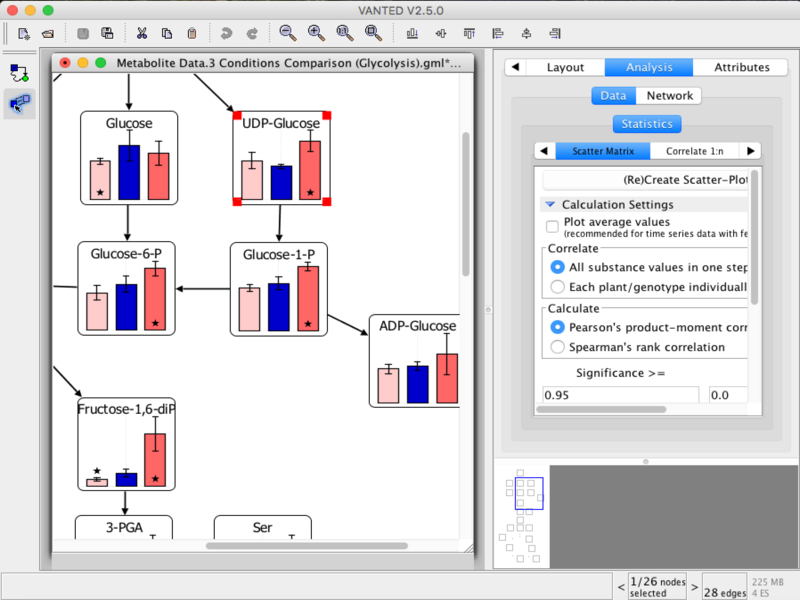
Vanted (Visualisation and Analysis of Networks conTaining Experimental Data) is a tool for the visual exploration and analysis of multimodal data in the context of biological networks. It helps to integrate and process data from different areas (such as transcriptomics, proteomics, metabolomics) and to present the results in a user-friendly way. Extensions based on Vanted address, for example, metabolic modelling (FBA-SimVis), an information system for experimental data (DBE2), network analysis methods (CentiLib), and spatial data investigations/image processing (Hive).
H. Rohn, A. Junker et al.: ‘Vanted v2: a framework for systems biology applications’ BMC Systems Biology 6: 139.1-13, 2012
B. H. Junker, C. Klukas and F. Schreiber: ‘Vanted: a system for advanced data analysis and visualization in the context of biological networks’ BMC Bioinformatics 7: 109.1-13, 2006.
TEAMwISE

TEAMwISE is a framework for collaborative analysis of geospatial-temporal movement data, for example, for collective behavior analysis. It provides different perspectives on the same data in collocated or remote setups, and supports the concurrent usage of several program instances. The implementation can be deployed in a variety of immersive environments, such as on tiled display walls or mobile VR devices. TEAMwISE allows analysts to investigate large and complex movement datasets in the context of the geographic environment it occurred in.
Available soon.
K. Klein, M. Aichem et al.: ‘TEAMwISE: Synchronised Immersive Environments for Exploration and Analysis of Movement Data’ (Proc. Visual Information Communication and Interaction - VINCI) 9.1-9.5, 2019.
Meta-All & MetaCrop
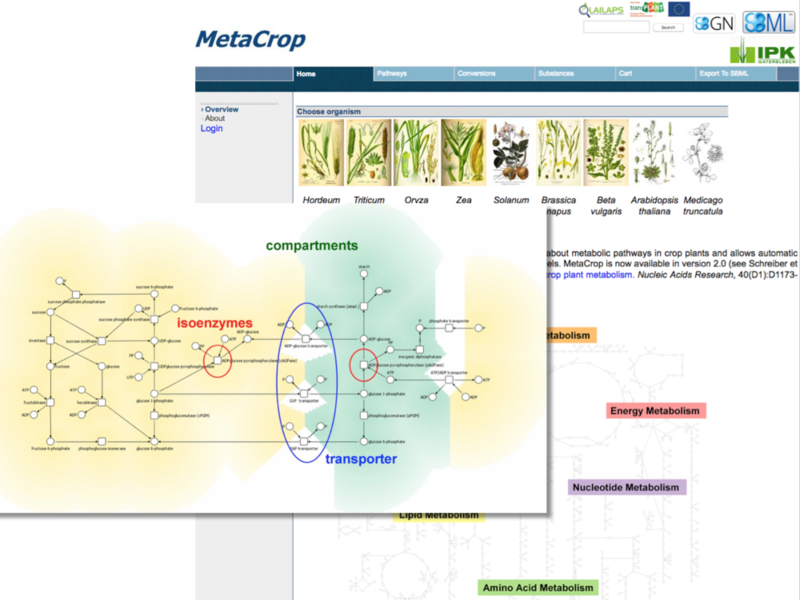
Meta-All is a database and information system for multi-level representation of metabolic processes, MetaCrop is a special instance of the system for detailed plant metabolism. MetaCrop is both a large collection of manually curated pathways and metabolic information in crop plants at different levels of detail (from overviews down to kinetic parameters) and a repository for metabolic models supporting standards such as SBML and SBGN.
F. Schreiber, C. Colmsee et al.: ‘MetaCrop 2.0: managing and exploring information about crop plant metabolism’ Nucleic Acids Research 40(1): D1173-D1177, 2012.
SBGN-ED
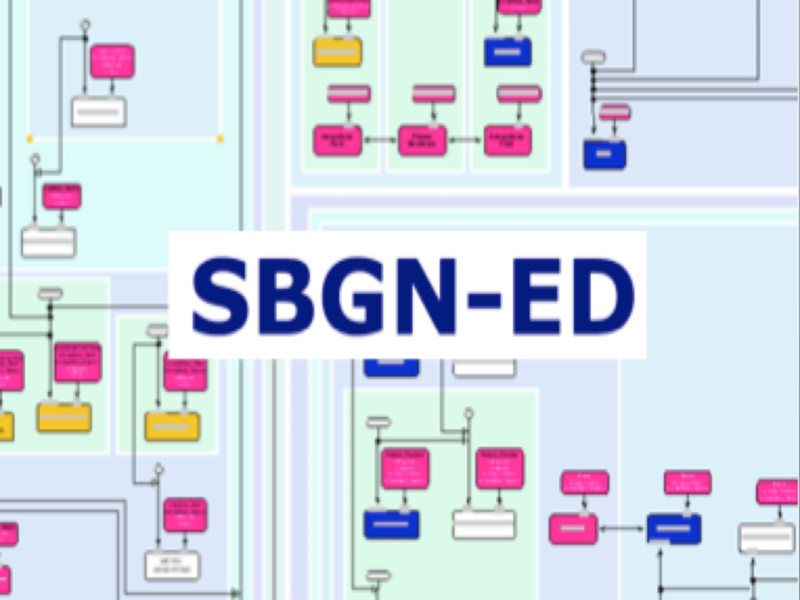
SBGN (Systems Biology Graphical Notation) is a standard for the graphical representation of biological networks and cellular systems. The software SBGN-ED supports editing, validation and interactive navigation of SBGN maps as well as mapping of data onto biological networks and cellular processes represented in SBGN, and their analysis.
T. Czauderna, C. Klukas and F. Schreiber: ‘Editing, validating, and translating of SBGN maps’ Bioinformatics 26(18): 2340-2341, 2010.
LMME
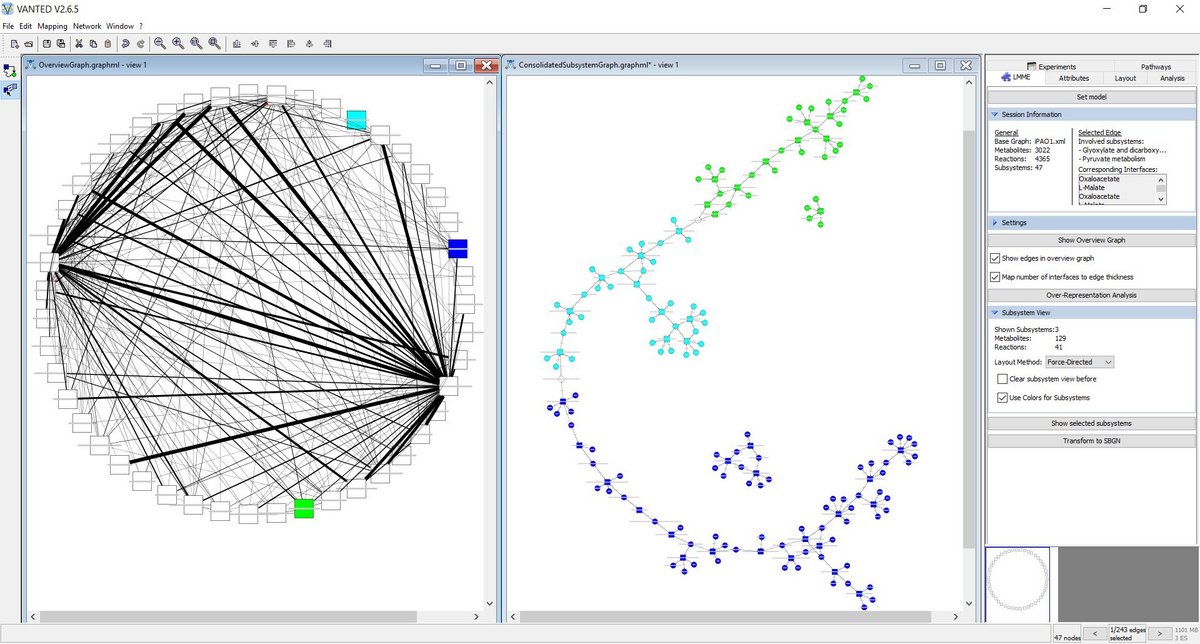
LMME (Large Metabolic Model Explorer), a Vanted Add-on, facilitates the exploration of large metabolic models based on decompositions of the underlying network into meaningful subsystems. It offers methods to hierarchically explore these networks and provides analysis techniques that may be applied successively.
www.cls.uni-konstanz.de/software/lmme
M. Aichem, T. Czauderna et al.: Visual Exploration of Large Metabolic Models. Bioinformatics, btab335 (2021).
CELLmicrocosmos
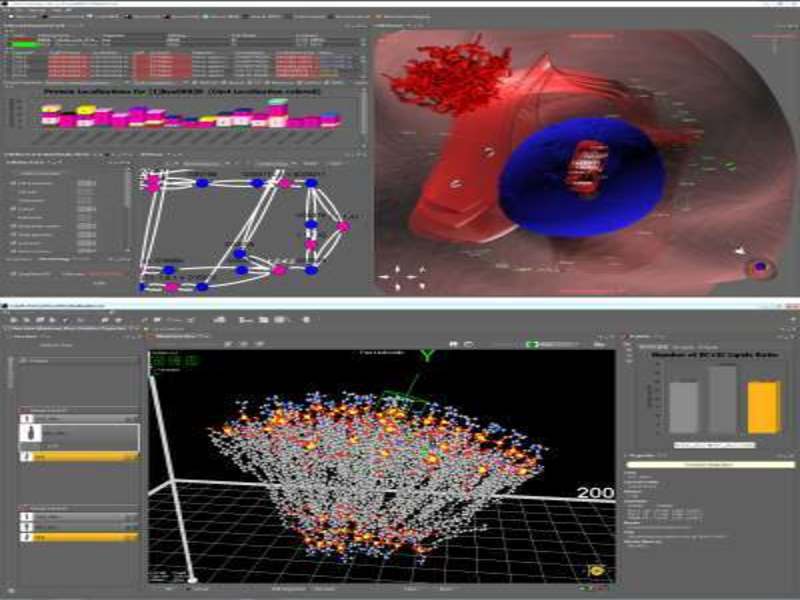
CELLmicrocosmos (short Cm) aims at the spatial analysis and interactive 3D visualisation of intracellular interactions and consists of different software applications such as Cm PathwayIntegration and Cm MembraneEditor. Cm PathwayIntegration supports the localisation and visualisation of biological pathways and other protein-/gene-related networks in the context of 3D cell models. Cm MembraneEditor generates models of molecular membranes and vesicular structures using PDB files ready for molecular dynamic simulations.
B. Sommer and F. Schreiber: ‘Integration and virtual reality exploration of biomedical data with CmPI and VANTED’ it – Information Technology 59(4): 181-190, 2017.
Contributions to Software Projects
ScaffoldHunter
Scaffold Hunter is a tool for the visual analysis of data from the life sciences, aiming at intuitive access to large and complex data sets that offers a variety of views and analysis methods. It originates from drug discovery and has evolved into a reusable open source platform for a wider range of applications.
Path2Models
Path2Models automatically generates mathematical models from biological pathway representations using a suite of freely available software. Computed models from over 2600 organisms encoded in SBML and SBGN are available through the BioModels Database.
OGDF & libSBGN
We contribute to open source libraries such as OGDF and libSBGN. OGDF is a self-contained C++ class library for the automatic layout of diagrams and offers sophisticated algorithms and data structures which can be used within your own applications or scientific projects. libSBGN is a software library for working with SBGN-ML files.
Optimas-DW
The Optimas Data Warehouse is a comprehensive data collection for maize and integrates data from different domains such as transcriptomics, metabolomics, ionomics, proteomics and phenomics. It provides interactive access to and analysis of the data.
Former Software Projects of the Research Group
HTPheno
Image analysis pipeline for high throughput plant phenotyping.
MAVisto, CentiBiN & CentiLib
Investigation of structural network properties (motifs, centralities, and global graph properties) especially for biological networks. Initially stand-alone programs, the newer developments for centralities and global graph properties are now part of the Vanted tool suite and further developed there.
BioPath
Representation, visualisation and exploration of biochemical pathways; an electronic version of the Boehringer pathway chart.
